About
MICCAI Workshop on Mesh Processing in Medical Image AnalysisIn conjunction with MICCAI 2013.
Nagoya, Japan
Nagoya University, Room IB013
September 26, 2013
Scope
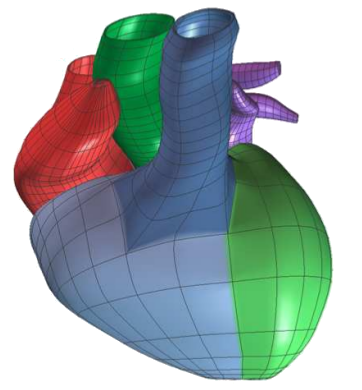 Many strategies for medical image analysis have been built on an
image analysis pipeline that starts with acquired image data,
performs filtering and processing, constructs geometric models of
important surfaces and structures, performs simulation, and finally
provides quantitative and visual analysis of the data. Within this
pipeline, geometry and shape are commonly represented as a mesh, or a
discretization of some domain into simpler computational elements such as
quads or triangles (representative of surface pieces) or tetrahedra and
hexahedra (representative of volumetric elements). This
image-to-mesh (I2M) step converts volumetric images into formats
that are more suitable for solving finite-element simulations, analyzing
critical structures, and performing boundary surface visualization tasks.
Current research in computational geometry, graphics hardware, and
computer graphics has produced methods to represent, extract, refine,
visualize and analyze both critical surfaces embedded in the 3D volumes,
such as interfaces between tissues, as well as volumetric regions, such as
organs.
Many strategies for medical image analysis have been built on an
image analysis pipeline that starts with acquired image data,
performs filtering and processing, constructs geometric models of
important surfaces and structures, performs simulation, and finally
provides quantitative and visual analysis of the data. Within this
pipeline, geometry and shape are commonly represented as a mesh, or a
discretization of some domain into simpler computational elements such as
quads or triangles (representative of surface pieces) or tetrahedra and
hexahedra (representative of volumetric elements). This
image-to-mesh (I2M) step converts volumetric images into formats
that are more suitable for solving finite-element simulations, analyzing
critical structures, and performing boundary surface visualization tasks.
Current research in computational geometry, graphics hardware, and
computer graphics has produced methods to represent, extract, refine,
visualize and analyze both critical surfaces embedded in the 3D volumes,
such as interfaces between tissues, as well as volumetric regions, such as
organs.
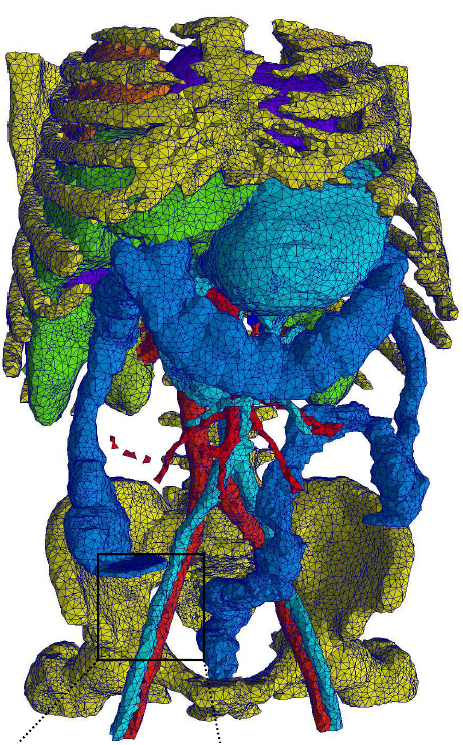 The workshop investigates the role meshes have with medical image
analysis and is broadly based on three overlapping themes:
The workshop investigates the role meshes have with medical image
analysis and is broadly based on three overlapping themes:
- Mesh processing,
- The I2M pipeline,
- Surface analysis and extraction.
Proceedings
Printed proceedings will be available at the workshop. However, they will not be published by Springer, as originally announced. Proceedings for MeshMed 2012 was published by Springer.Best paper prize
The best paper prize for MeshMed 2013 went to Sergio Vera and his coworkers for the paper: Volumetric Anatomical Parameterization and Meshing for Inter-patient Liver Coordinate System Definition. Congratulations!Registration
Registration for the workshop is handled trough the main MICCAI conference.Organisers

2013 Chairs
- Joshua A. Levine, SCI Institute, University of Utah, USA
- Rasmus R. Paulsen, Technical University of Denmark, Denmark
- Yongjie Zhang, Carnegie Mellon University, USA
Steering Comittee
- Hervé Delingette, Asclepios, INRIA Sophia-Antipolis, France
- Nikos P. Chrisochoides, Old Dominion University, USA
- Sylvain Prima, IRISA / INRIA Rennes, France
Sponsors
MeshMed is supported by the HEAR-EU project funded from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement number 304857.Contact
|
Rasmus R. Paulsen Associate Professor Applied Mathematics and Computer Science Technical University of Denmark Matematiktorvet Building 324, office 110 DK-2800 Kgs. Lyngby Denmark Phone: +45 4525 3423 Fax: +45 4588 1397 http://www.imm.dtu.dk/~rrp rrp at imm dot dtu dot dk |
Joshua A. Levine Assistant Professor School of Computing Visual Computing Division Clemson University 317 McAdams Hall Clemson, SC 29634 USA Phone: +1-864-656-0537 Fax: http://people.cs.clemson.edu/~levinej/ levinej at clemson dot edu |
Yongjie (Jessica) Zhang Associate Professor Department of Mechanical Engineering Carnegie Mellon University 303 Scaife Hall 5000 Forbes Avenue Pittsburgh, PA 15213 USA Phone: +1-412-268-5332 Fax: +1-412-268-3348 http://www.andrew.cmu.edu/ user/jessicaz/ jessicaz at andrew dot cmu dot edu |



